Molecular casings makes DNA-computing in blood viable by protecting it from enzyme attack
DNA molecules can potentially be reprogrammed to help do useful things, like detecting diseases and releasing drugs. But until now this so-called DNA computing was impossible in blood, as human enzymes degrade the molecules almost instantly. Biomedical Engineer Tom de Greef from Eindhoven University of Technology together with researchers from Radboud University, University of Bristol and Microsoft Research have solved this issue by creating a protective molecular casings, in which they created functioning DNA-based computational circuits. This compartmentalization approach has a nice bonus: it also increases the computing speed. The results are published today in Nature Nanotechnology.
Desktop computers use a series of logic gates to transform electric inputs into outputs. In a similar manner, molecular computers made from DNA use programmable interactions between DNA strands to transform DNA inputs into outputs. DNA computers can be programmed to perform complex calculations on molecular data without human intervention.
In a study published today in Nature Nanotechnology, a team led by Tom de Greef, Associate Professor in Synthetic Biology at the Department of Biomedical Engineering of TU/e, and by Professor Stephen Mann from the University of Bristol’s School of Chemistry, presents a new approach for DNA-based computers. This approach uses communities of capsules (‘proteinosomes’). These capsules contain a diversity of DNA molecules that together can be used for molecular sensing and computation, with potential applications in in-vitro diagnostics and smart therapeutics.

Faster, modular, effective
DNA computers are inherently slow and poorly scalable because they operate in a ‘chemical soup’ where they rely on random diffusion to interact with each other and execute a computational step. De Greef: “With the introduction of compartmentalization, we increase the concentration of DNA molecules inside the capsules and, thus, the computing speed. Also, compartmentalization enables a modular design of the computational circuits.”
Indestructible
One of the long-standing goals of nanotechnology is to create autonomous molecular machines that can operate in harsh biological environments. “To date”, explains De Greef, “DNA-based computers still cannot be used in biological relevant environments as enzymes present in blood serum would destroy the DNA strands responsible for computations.” In the approach proposed in this study, the encapsulation of DNA molecules inside proteinosomes makes them less vulnerable to digestion by enzymes. Thus, their lifetime in blood serum is greatly increased, opening the way to the development of real, cell-like autonomous systems operating in physiological conditions.
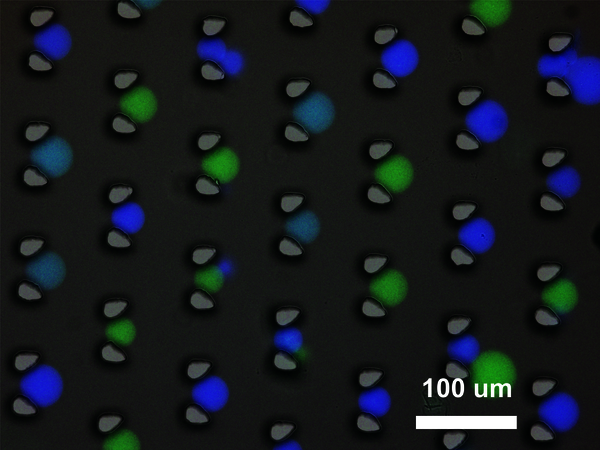
Bi-directional communication
Living cells communicate by secreting diffusible signaling molecules that activate key molecular processes in neighboring cells. These intercellular communication is often bi-directional and include both positive and negative regulatory interactions. De Greef: “With our platform, we mimicked intercellular communication occurring in living systems by using two communities of artificial cells. For example, we showed that, when an input DNA strand enters and activates the first community, the secreted signal activates the second community. The latter responds secreting an inhibitor which deactivates back the first community.”
New applications in the medical field
This new approach lays the groundwork for using protocell communication platforms to bring embedded molecular control circuits closer to practical applications in the medical field. Currently, researchers at the TU/e and Microsoft are testing the compartmentalized DNA circuits to detect and classify diseases.
‘DNA-based communication in populations of synthetic protocells’ by A. Joesaar et al. is published today in Nature Nanotechnology (DOI: 1038/s41565-019-0399-9).